Oncogenesis, the pathogenesis of neoplasm, involves either gene itself or a product, i.e. protein.
Oncogenes
Oncogenes are the genes that control autonomous cell growth in cancer cells. Oncogenes arise from proto-oncogenes.
A proto-oncogene is a normal gene that can become an oncogene due to mutations.
Functions of oncogenes:
- Over-expression of proto-oncogene required for the production of growth factors. The transformed cell will produce growth factor in autocrine pattern.
- Oncogenes encode for growth factor receptors. These receptors will be activated even without binding with growth factors.
- They act as signal transducing proteins i.e. they receive signals from outside the cells and transmit them to cell nucleus. RAS gene produces RAS protein for this purpose in many cancers.
- Act as transcription factors and have role in cell cycle regulation i.e. MYC gene.
- Act as cyclins and cyclin dependent kinases to control cell cycle.
Cell cycle check points
- At G1/S transition
- At G2/M transition
These check points are under genetic control.
Gate keeper genes are the group of gees that prevent cell cycle progression till DNA damage is repaired. Any alteration in gene leads to tumor formation.
Retinoblastoma gene (RB gene)
First tumor suppressor gene discovered in retinoblastoma.
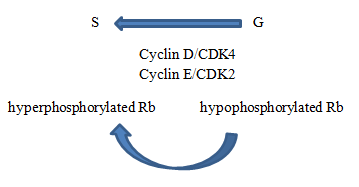
Active state of Rb gene
In hypophosphorylated state, RB is active and acts as tumor suppressor by inhibiting cell cycle progression.
Role of cyclin dependent kinases (CDK)
When it is time for cell to enter S phase, complexes of CDK and cyclins phosphorylate Rb, inhibiting its activity. Molecular brakes from cell cycle are released.
- G0/G1 non-phosphorylated
- G1/S phosphorylated by cyclin/CDK
Control of G1 phase in normal cell
Initiation of DNA replication requires normal activity of cyclin E.
Normal expression of cyclin E is dependent on E2F family of transcription factors (targets of RB genes).
- RB in active form binds to and inhibits E2F family of transcription factors and prevents transcription of cyclin E.
- RB also controls stability of cell cycle inhibitor p27.
Mitogenic signals phosphorylate RB and brakes are released –cell will enter into S phase.
RB gene mutations
Due to RB gene mutations, brakes on cell cycle are released and cell moves through cell cycle.
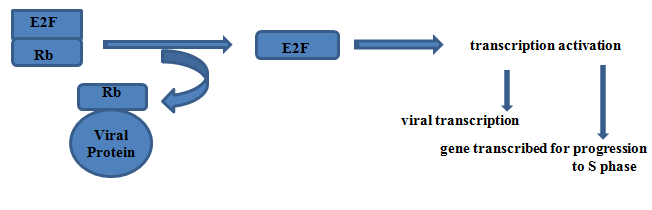
Targets of RB
The targets of RB genes are the E2F group of transcription factors, released E2F prompts cell to enter S phase.
RB pocket
Pocket domain of retinoblastoma (RB) tumor suppressor is central to Rb function, mutations are found in this localized area.
Pathways for RB gene regulation
- Growth factors
- TGF-beta
- P53
- CDK inhibitors
Associated tumors
Deletion or inactivation of RB gene plays an essential role in:
- Retinoblastoma
- Osteosarcomas
RB inactivation is also found in other tumors, including:
- Sarcomas
- Small cell carcinoma of lung
- Carcinoma of breast
- Carcinoma of Bladder
- Carcinoma of Prostate
Two hit hypothesis for retinoblastoma
Predisposing conditions may be inherited or acquired. 2nd hit after birth (radiations, viruses) leads to the formation of mutated RB gene, as in retinoblastoma.
Inactivation of Rb protein by binding with viral protein
P53 gene –Guardian of Genone
Located on chromosome 17p13.1
Functions
1. Cell cycle arrest
P53 acts as gate keeper, it senses DNA damage and prevents propagation of genetically damaged cells.
2. DNA repair
This gene assists in DNA repair. It acts as a transcription factor.
3. Initiation of apoptosis
Cells with unrepairable DNA are directed to apoptosis by p53 gene.
4. Controls other genes
p53 also mediates cell cycle arrest and apoptosis.
How p53 prevents neoplastic transformation?
By three mechanisms:
- Activation of temporary cell cycle arrest
- Permanent cell cycle arrest (senescence)
- Triggering of programmed cell death (apoptosis)
1. Activation of temporary cell cycle arrest
- In healthy cell p53 levels are low with half life of 20 minutes.
- MDM2 proteins control these levels by removing p53 from nucleus and degradation in cytoplasm.
- If cell is under stress and there is DNA damage, p53 will be phosphorylated and acetylated.
- MDM2 cannot bind with this active form of p53 and degradation is prevented.
- Activated p53 transcript a number of genes for DNA repair
- If DNA damage is repaired successfully, p53 upregulates transcription of MDM2, leading to own destruction and thus releasing the cell cycle block.
2. Permanent Cell Cycle Arrest
- If DNA damage is not repaired, cell will enter into p53 induced senescence.
- Senescence is a permanent cell cycle arrest characterized by specific changes in morphology and gene expression of cell.
- It requires activation of p53, Rb and other related genes. Like all p53 responses, senescence is stimulated by a variety of stresses, like hypoxia.
- But still exact mechanism is not known as yet.
3. Triggering of programmed cell death (apoptosis)
- It is ultimate protective mechanism against neoplastic transformation.
- If DNA damage is not repaired, enough p53 accumulates to transcript pro-apoptotic genes and cell dies.
Inactivation of p53 by binding with Viral Proteins
Viral inactivation of p53 function
Tumor association of p53
Over half of human malignant cells show mutated p53 gene.
Most common examples are of:
- Lungs
- Colon
- Carcinoma of breast
Therapeutic implication of p53
- Based on property of p53 to control apoptosis in response to DNA damage.
- Irradiation and chemotherapeutic agents mediate effect by inducing DNA damage and subsequent apoptosis.
- By lethal DNA damage property of p53 is utilized.
Tumors that retain normal p53 are more likely to respond to such therapy as:
- ALL (acute lymphoblastic lymphoma)
- Teratocarcinoma
 howMed Know Yourself
howMed Know Yourself